Introduction to pandas#
Note
This material is mostly adapted from the following resources:

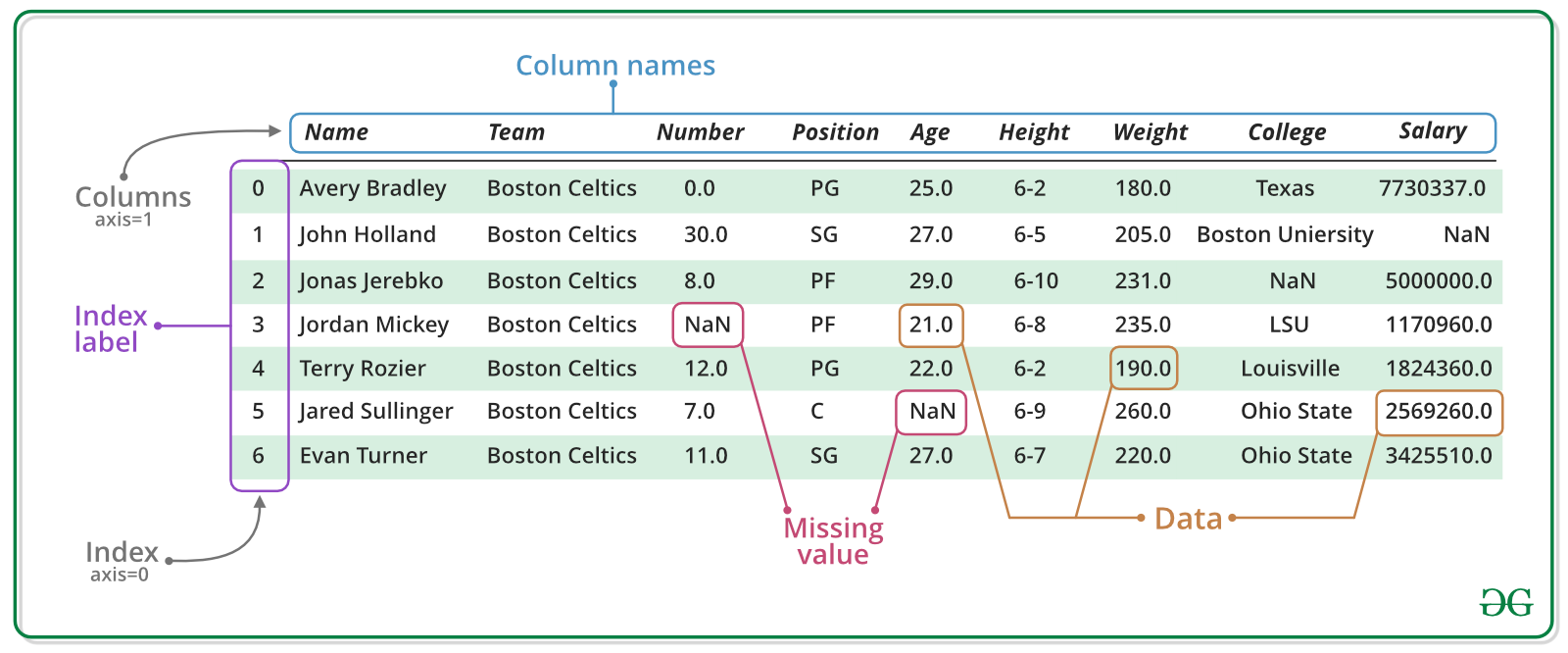
Pandas is a an open source library providing Excel-like tables in Python. It offers functionality for efficiently reading, writing, and processing data such as sorting, filtering, aggregating, and visualizing. Moreover, it provides tools for handling missing data and time series data.
Note
Documentation for this package is available at https://pandas.pydata.org/docs/.
Note
If you have not yet set up Python on your computer, you can execute this tutorial in your browser via Google Colab. Click on the rocket in the top right corner and launch “Colab”. If that doesn’t work download the .ipynb file and import it in Google Colab.
Then install pandas and numpy by executing the following command in a Jupyter cell at the top of the notebook.
!pip install pandas numpy
import pandas as pd
import numpy as np
The Series#
A Series represents a one-dimensional array of data. The main difference between a Series and numpy array is that a Series has an index. The index contains the labels that we use to access the data. It is actually quite similar to a Python dictionary, where each value is associated with a key.
There are many ways to create a Series, but the core constructor is pd.Series() which can process a dictionary to create a Series.
Note
The data used below is from Wikipedia’s List of power stations in Germany.
dictionary = {
"Neckarwestheim": 1269,
"Isar 2": 1365,
"Emsland": 1290,
}
s = pd.Series(dictionary)
s
Neckarwestheim 1269
Isar 2 1365
Emsland 1290
dtype: int64
dictionary
{'Neckarwestheim': 1269, 'Isar 2': 1365, 'Emsland': 1290}
Arithmetic operations and most numpy functions can be applied to pd.Series.
An important point is that the Series keep their index during such operations.
np.log(s) / s**0.5
Neckarwestheim 0.200600
Isar 2 0.195391
Emsland 0.199418
dtype: float64
We can access the underlying index object if we need to:
s.index
Index(['Neckarwestheim', 'Isar 2', 'Emsland'], dtype='object')
We can get values back out using the index via the .loc attribute
s.loc["Isar 2"]
np.int64(1365)
Or by raw position using .iloc
s.iloc[2]
np.int64(1290)
We can pass a list or array to loc to get multiple rows back:
s.loc[["Neckarwestheim", "Emsland"]]
Neckarwestheim 1269
Emsland 1290
dtype: int64
And we can even use so-called slicing notation (:) to get ranges of rows:
s.loc["Neckarwestheim":"Emsland"]
Neckarwestheim 1269
Isar 2 1365
Emsland 1290
dtype: int64
s.iloc[:2]
Neckarwestheim 1269
Isar 2 1365
dtype: int64
If we need to, we can always get the raw data back out as well
type(s.values) # a numpy array
numpy.ndarray
The DataFrame#
There is a lot more to a pandas.Series, but they are limit to a single column. A more broadly useful Pandas data structure is the DataFrame. pandas.DataFrame is a collection of series that share the same index. It’s a lot like a table in a spreadsheet.
The core constructor is pd.DataFrame(), which can be used like this using a dictionary of lists:
data = {
"capacity": [1269, 1365, 1290], # MW
"type": ["PWR", "PWR", "PWR"],
"start_year": [1989, 1988, 1988],
"end_year": [np.nan, np.nan, np.nan],
}
df = pd.DataFrame(data, index=["Neckarwestheim", "Isar 2", "Emsland"])
df
| capacity | type | start_year | end_year | |
|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | NaN |
| Isar 2 | 1365 | PWR | 1988 | NaN |
| Emsland | 1290 | PWR | 1988 | NaN |
We can also switch columns and rows very easily using the .T (transpose) attribute:
df
| capacity | type | start_year | end_year | |
|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | NaN |
| Isar 2 | 1365 | PWR | 1988 | NaN |
| Emsland | 1290 | PWR | 1988 | NaN |
A wide range of statistical functions are available on both Series and DataFrames.
df.min()
capacity 1269
type PWR
start_year 1988
end_year NaN
dtype: object
df.mean(numeric_only=True)
capacity 1308.000000
start_year 1988.333333
end_year NaN
dtype: float64
df.describe()
| capacity | start_year | end_year | |
|---|---|---|---|
| count | 3.000000 | 3.000000 | 0.0 |
| mean | 1308.000000 | 1988.333333 | NaN |
| std | 50.467812 | 0.577350 | NaN |
| min | 1269.000000 | 1988.000000 | NaN |
| 25% | 1279.500000 | 1988.000000 | NaN |
| 50% | 1290.000000 | 1988.000000 | NaN |
| 75% | 1327.500000 | 1988.500000 | NaN |
| max | 1365.000000 | 1989.000000 | NaN |
We can get a single column as a Series using python’s getitem syntax on the DataFrame object.
df["capacity"]
Neckarwestheim 1269
Isar 2 1365
Emsland 1290
Name: capacity, dtype: int64
…or using attribute syntax.
df.end_year
Neckarwestheim NaN
Isar 2 NaN
Emsland NaN
Name: end_year, dtype: float64
Indexing works very similar to series
df.loc["Emsland"]
capacity 1290
type PWR
start_year 1988
end_year NaN
Name: Emsland, dtype: object
df.iloc[2]
capacity 1290
type PWR
start_year 1988
end_year NaN
Name: Emsland, dtype: object
But we can also specify the column(s) and row(s) we want to access
df.loc["Emsland", "start_year"]
np.int64(1988)
df.loc[["Emsland", "Neckarwestheim"], ["start_year", "end_year"]]
| start_year | end_year | |
|---|---|---|
| Emsland | 1988 | NaN |
| Neckarwestheim | 1989 | NaN |
Mathematical operations work as well, either on the whole DataFrame or on specific columns, the result of which can be assigned to a new column:
df.capacity * 0.8
Neckarwestheim 1015.2
Isar 2 1092.0
Emsland 1032.0
Name: capacity, dtype: float64
df["reduced_capacity"] = df.capacity * 0.8
df
| capacity | type | start_year | end_year | reduced_capacity | |
|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | NaN | 1015.2 |
| Isar 2 | 1365 | PWR | 1988 | NaN | 1092.0 |
| Emsland | 1290 | PWR | 1988 | NaN | 1032.0 |
Cleaning Data#
We can also remove columns or rows from a DataFrame:
df.drop("reduced_capacity", axis="columns")
df
| capacity | type | start_year | end_year | reduced_capacity | |
|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | NaN | 1015.2 |
| Isar 2 | 1365 | PWR | 1988 | NaN | 1092.0 |
| Emsland | 1290 | PWR | 1988 | NaN | 1032.0 |
We can update the variable df by either overwriting df or passing an inplace keyword:
df = df.drop("reduced_capacity", axis="columns")
df
| capacity | type | start_year | end_year | |
|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | NaN |
| Isar 2 | 1365 | PWR | 1988 | NaN |
| Emsland | 1290 | PWR | 1988 | NaN |
We can also drop columns with only NaN values
df.dropna(axis=1, how="any")
| capacity | type | start_year | |
|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 |
| Isar 2 | 1365 | PWR | 1988 |
| Emsland | 1290 | PWR | 1988 |
Or fill it up with default fallback data:
df.fillna(2023)
df
| capacity | type | start_year | end_year | |
|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | NaN |
| Isar 2 | 1365 | PWR | 1988 | NaN |
| Emsland | 1290 | PWR | 1988 | NaN |
Say, we already have one value for end_year and want to fill up the missing data. We can use forward fill (ffill) or backward fill (bfill):
df.loc["Emsland", "end_year"] = 2023
df.loc["Neckarwestheim", "end_year"] = 2026
df
| capacity | type | start_year | end_year | |
|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 |
| Isar 2 | 1365 | PWR | 1988 | NaN |
| Emsland | 1290 | PWR | 1988 | 2023.0 |
df["end_year"] = df["end_year"].ffill()
df
df["x"] = np.nan
df["y"] = np.nan
df
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 | NaN | NaN |
| Isar 2 | 1365 | PWR | 1988 | 2026.0 | NaN | NaN |
| Emsland | 1290 | PWR | 1988 | 2023.0 | NaN | NaN |
Sometimes it can be useful to rename columns:
df.rename(columns=dict(x="lat", y="lon"))
| capacity | type | start_year | end_year | lat | lon | |
|---|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 | NaN | NaN |
| Isar 2 | 1365 | PWR | 1988 | 2026.0 | NaN | NaN |
| Emsland | 1290 | PWR | 1988 | 2023.0 | NaN | NaN |
Sometimes it can be useful to replace values:
df.replace({"PWR": "Pressurized water reactor"})
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Neckarwestheim | 1269 | Pressurized water reactor | 1989 | 2026.0 | NaN | NaN |
| Isar 2 | 1365 | Pressurized water reactor | 1988 | 2026.0 | NaN | NaN |
| Emsland | 1290 | Pressurized water reactor | 1988 | 2023.0 | NaN | NaN |
df
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 | NaN | NaN |
| Isar 2 | 1365 | PWR | 1988 | 2026.0 | NaN | NaN |
| Emsland | 1290 | PWR | 1988 | 2023.0 | NaN | NaN |
In many cases, we want to modify values in a dataframe based on some rule. To modify values, we need to use .loc or .iloc. It can be use to set a specific value or a set of values based on their index and column labels:
df.loc["Isar 2", "start_year"] = 2000
df.loc["Emsland", "capacity"] += 10
df
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 | NaN | NaN |
| Isar 2 | 1365 | PWR | 2000 | 2026.0 | NaN | NaN |
| Emsland | 1300 | PWR | 1988 | 2023.0 | NaN | NaN |
It can even be a completely new column:
operational = ["Neckarwestheim", "Isar 2", "Emsland"]
df.loc[operational, "y"] = [49.04, 48.61, 52.47]
df
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 | NaN | 49.04 |
| Isar 2 | 1365 | PWR | 2000 | 2026.0 | NaN | 48.61 |
| Emsland | 1300 | PWR | 1988 | 2023.0 | NaN | 52.47 |
Combining Datasets#
Pandas supports a wide range of methods for merging different datasets. These are described extensively in the documentation. Here we just give a few examples.
data = {
"capacity": [1288, 1360, 1326], # MW
"type": ["BWR", "PWR", "PWR"],
"start_year": [1985, 1985, 1986],
"end_year": [2021, 2021, 2021],
"x": [10.40, 9.41, 9.35],
"y": [48.51, 52.03, 53.85],
}
df2 = pd.DataFrame(data, index=["Gundremmingen", "Grohnde", "Brokdorf"])
df2
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Gundremmingen | 1288 | BWR | 1985 | 2021 | 10.40 | 48.51 |
| Grohnde | 1360 | PWR | 1985 | 2021 | 9.41 | 52.03 |
| Brokdorf | 1326 | PWR | 1986 | 2021 | 9.35 | 53.85 |
We can now add this additional data to the df object
df = pd.concat([df, df2])
df
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 | NaN | 49.04 |
| Isar 2 | 1365 | PWR | 2000 | 2026.0 | NaN | 48.61 |
| Emsland | 1300 | PWR | 1988 | 2023.0 | NaN | 52.47 |
| Gundremmingen | 1288 | BWR | 1985 | 2021.0 | 10.40 | 48.51 |
| Grohnde | 1360 | PWR | 1985 | 2021.0 | 9.41 | 52.03 |
| Brokdorf | 1326 | PWR | 1986 | 2021.0 | 9.35 | 53.85 |
Sorting & Filtering Data#
We can also sort the entries in dataframes, e.g. alphabetically by index or numerically by column values
df.sort_index()
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Brokdorf | 1326 | PWR | 1986 | 2021.0 | 9.35 | 53.85 |
| Emsland | 1300 | PWR | 1988 | 2023.0 | NaN | 52.47 |
| Grohnde | 1360 | PWR | 1985 | 2021.0 | 9.41 | 52.03 |
| Gundremmingen | 1288 | BWR | 1985 | 2021.0 | 10.40 | 48.51 |
| Isar 2 | 1365 | PWR | 2000 | 2026.0 | NaN | 48.61 |
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 | NaN | 49.04 |
df.sort_values(by="end_year", ascending=False)
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 | NaN | 49.04 |
| Isar 2 | 1365 | PWR | 2000 | 2026.0 | NaN | 48.61 |
| Emsland | 1300 | PWR | 1988 | 2023.0 | NaN | 52.47 |
| Gundremmingen | 1288 | BWR | 1985 | 2021.0 | 10.40 | 48.51 |
| Grohnde | 1360 | PWR | 1985 | 2021.0 | 9.41 | 52.03 |
| Brokdorf | 1326 | PWR | 1986 | 2021.0 | 9.35 | 53.85 |
We can also filter a DataFrame using a boolean series obtained from a condition. This is very useful to build subsets of the DataFrame.
df.capacity > 1300
Neckarwestheim False
Isar 2 True
Emsland False
Gundremmingen False
Grohnde True
Brokdorf True
Name: capacity, dtype: bool
df[df.capacity > 1300]
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Isar 2 | 1365 | PWR | 2000 | 2026.0 | NaN | 48.61 |
| Grohnde | 1360 | PWR | 1985 | 2021.0 | 9.41 | 52.03 |
| Brokdorf | 1326 | PWR | 1986 | 2021.0 | 9.35 | 53.85 |
We can also combine multiple conditions, but we need to wrap the conditions with brackets!
df[(df.capacity > 1300) & (df.start_year >= 1988)]
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Isar 2 | 1365 | PWR | 2000 | 2026.0 | NaN | 48.61 |
Or we make SQL-like queries:
df.query("start_year == 1988")
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Emsland | 1300 | PWR | 1988 | 2023.0 | NaN | 52.47 |
threshold = 1300
df.query("start_year == 1988 and capacity > @threshold")
| capacity | type | start_year | end_year | x | y |
|---|
df
| capacity | type | start_year | end_year | x | y | |
|---|---|---|---|---|---|---|
| Neckarwestheim | 1269 | PWR | 1989 | 2026.0 | NaN | 49.04 |
| Isar 2 | 1365 | PWR | 2000 | 2026.0 | NaN | 48.61 |
| Emsland | 1300 | PWR | 1988 | 2023.0 | NaN | 52.47 |
| Gundremmingen | 1288 | BWR | 1985 | 2021.0 | 10.40 | 48.51 |
| Grohnde | 1360 | PWR | 1985 | 2021.0 | 9.41 | 52.03 |
| Brokdorf | 1326 | PWR | 1986 | 2021.0 | 9.35 | 53.85 |
Applying Functions#
Sometimes it can be useful to apply a function to all values of a column/row. For instance, we might be interested in normalised capacities relative to the largest nuclear power plant:
def normalise(s):
return s / df.capacity.max()
df.capacity.apply(normalise)
Neckarwestheim 0.929670
Isar 2 1.000000
Emsland 0.952381
Gundremmingen 0.943590
Grohnde 0.996337
Brokdorf 0.971429
Name: capacity, dtype: float64
For simple functions, there’s often an easier alternative:
def normalise(s: float):
# adsfjielfwa
return s / df.capacity.max()
df.capacity.apply(normalise)
Neckarwestheim 0.929670
Isar 2 1.000000
Emsland 0.952381
Gundremmingen 0.943590
Grohnde 0.996337
Brokdorf 0.971429
Name: capacity, dtype: float64
But the .apply() function often gives you more flexibility.
Plotting#
DataFrames have all kinds of useful plotting built in.
Note
Note, that we do not even have to import matplotlib for this. It is used under the hood by pandas to create the plots.
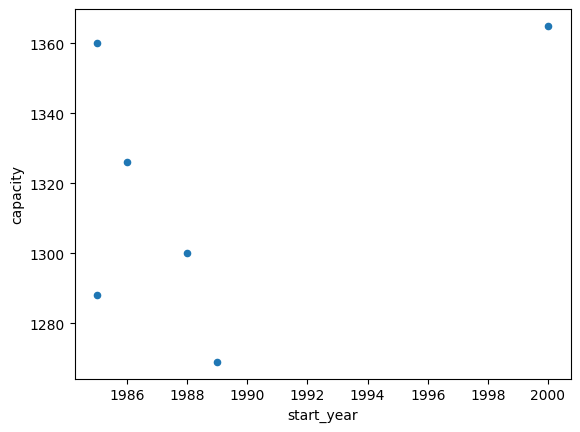
df.plot(kind="scatter", x="start_year", y="capacity")
<Axes: xlabel='start_year', ylabel='capacity'>
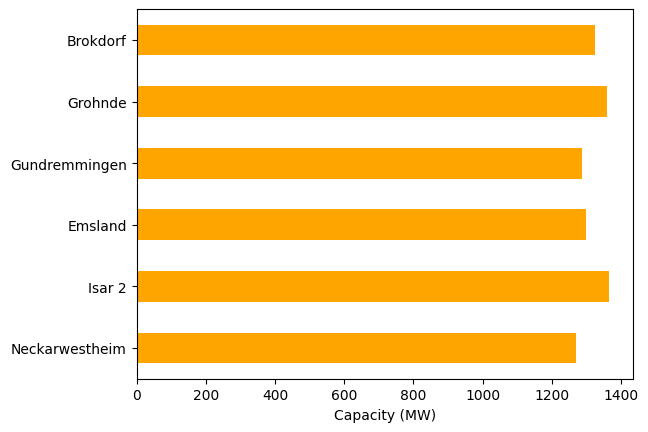
df.capacity.plot.barh(color="orange")
import matplotlib.pyplot as plt
plt.xlabel("Capacity (MW)")
Text(0.5, 0, 'Capacity (MW)')
Time Indexes#
Indexes are very powerful. They are a big part of why Pandas is so useful. There are different indices for different types of data. Time Indexes are especially great when handling time-dependent data.
time = pd.date_range(start="2021-01-01", end="2023-01-01", freq="D")
time.dayofyear
Index([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
...
357, 358, 359, 360, 361, 362, 363, 364, 365, 1],
dtype='int32', length=731)
values = np.sin(2 * np.pi * time.dayofyear / 365)
values
Index([ 0.017213356155834685, 0.03442161162274574, 0.051619667223253764,
0.06880242680231986, 0.08596479873744647, 0.10310169744743485,
0.1202080448993527, 0.13727877211326478, 0.15430882066428117,
0.1712931441814776,
...
-0.13727877211326517, -0.12020804489935275, -0.10310169744743544,
-0.0859647987374467, -0.06880242680232064, -0.05161966722325418,
-0.034421611622745804, -0.01721335615583528, 6.432490598706546e-16,
0.017213356155834685],
dtype='float64', length=731)
values = np.sin(2 * np.pi * time.dayofyear / 365)
ts = pd.Series(values, index=time)
ts
2021-01-01 1.721336e-02
2021-01-02 3.442161e-02
2021-01-03 5.161967e-02
2021-01-04 6.880243e-02
2021-01-05 8.596480e-02
...
2022-12-28 -5.161967e-02
2022-12-29 -3.442161e-02
2022-12-30 -1.721336e-02
2022-12-31 6.432491e-16
2023-01-01 1.721336e-02
Freq: D, Length: 731, dtype: float64
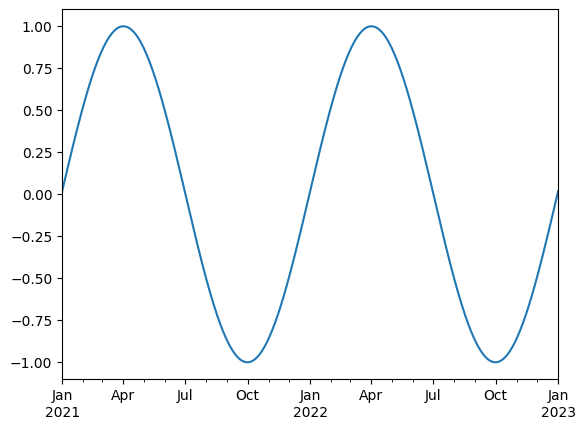
ts.plot()
<Axes: >
We can use Python’s slicing notation inside .loc to select a date range.
ts.loc["2021-01-01":"2021-07-01"].plot()
<Axes: >
ts.loc["2021-05"].plot()
<Axes: >
The pd.TimeIndex object has lots of useful attributes
ts.index.month
Index([ 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
...
12, 12, 12, 12, 12, 12, 12, 12, 12, 1],
dtype='int32', length=731)
ts.index.weekday
Index([4, 5, 6, 0, 1, 2, 3, 4, 5, 6,
...
4, 5, 6, 0, 1, 2, 3, 4, 5, 6],
dtype='int32', length=731)
Another common operation is to change the resolution of a dataset by resampling in time. Pandas exposes this through the .resample() function. The resample periods are specified using pandas offset index syntax.
Below, we resample the dataset by taking the mean over each month.
ts.resample("12h").mean().interpolate()
2021-01-01 00:00:00 1.721336e-02
2021-01-01 12:00:00 2.581748e-02
2021-01-02 00:00:00 3.442161e-02
2021-01-02 12:00:00 4.302064e-02
2021-01-03 00:00:00 5.161967e-02
...
2022-12-30 00:00:00 -1.721336e-02
2022-12-30 12:00:00 -8.606678e-03
2022-12-31 00:00:00 6.432491e-16
2022-12-31 12:00:00 8.606678e-03
2023-01-01 00:00:00 1.721336e-02
Freq: 12h, Length: 1461, dtype: float64
ts.resample("ME").mean().plot()
<Axes: >
Reading and Writing Files#
To read data into pandas, we can use for instance the pd.read_csv() function. This function is quite powerful and complex with many different settings. You can use it to extract data from almost any text file.
The pd.read_csv() function can take a path to a local file as an input, or even a hyperlink to an online text file.
Let’s import a slightly larger dataset about the power plant fleet in Europe_
fn = "https://raw.githubusercontent.com/PyPSA/powerplantmatching/master/powerplants.csv"
# fn = "powerplants(4).csv"
df = pd.read_csv(fn, index_col=0)
df = pd.read_csv(fn, index_col=0)
df.iloc[:5, :10]
| Name | Fueltype | Technology | Set | Country | Capacity | Efficiency | DateIn | DateRetrofit | DateOut | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 0 | Kernkraftwerk Emsland | Nuclear | Steam Turbine | PP | Germany | 1336.0 | 0.33 | 1988.0 | 1988.0 | 2023.0 |
| 1 | Brokdorf | Nuclear | Steam Turbine | PP | Germany | 1410.0 | 0.33 | 1986.0 | 1986.0 | 2021.0 |
| 2 | Borssele | Hard Coal | Steam Turbine | PP | Netherlands | 485.0 | NaN | 1973.0 | NaN | 2034.0 |
| 3 | Gemeinschaftskernkraftwerk Neckarwestheim | Nuclear | Steam Turbine | PP | Germany | 1310.0 | 0.33 | 1976.0 | 1989.0 | 2023.0 |
| 4 | Isar | Nuclear | Steam Turbine | PP | Germany | 1410.0 | 0.33 | 1979.0 | 1988.0 | 2023.0 |
df.info()
<class 'pandas.core.frame.DataFrame'>
Index: 29565 entries, 0 to 29862
Data columns (total 18 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Name 29565 non-null object
1 Fueltype 29565 non-null object
2 Technology 21661 non-null object
3 Set 29565 non-null object
4 Country 29565 non-null object
5 Capacity 29565 non-null float64
6 Efficiency 541 non-null float64
7 DateIn 23772 non-null float64
8 DateRetrofit 2436 non-null float64
9 DateOut 932 non-null float64
10 lat 29565 non-null float64
11 lon 29565 non-null float64
12 Duration 614 non-null float64
13 Volume_Mm3 29565 non-null float64
14 DamHeight_m 29565 non-null float64
15 StorageCapacity_MWh 29565 non-null float64
16 EIC 29565 non-null object
17 projectID 29565 non-null object
dtypes: float64(11), object(7)
memory usage: 4.3+ MB
df.describe()
| Capacity | Efficiency | DateIn | DateRetrofit | DateOut | lat | lon | Duration | Volume_Mm3 | DamHeight_m | StorageCapacity_MWh | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 29565.000000 | 541.000000 | 23772.000000 | 2436.000000 | 932.000000 | 29565.000000 | 29565.000000 | 614.000000 | 29565.000000 | 29565.000000 | 29565.000000 |
| mean | 46.318929 | 0.480259 | 2005.777764 | 1988.509442 | 2020.609442 | 49.636627 | 9.010803 | 1290.093123 | 2.889983 | 6.463436 | 355.629403 |
| std | 198.176762 | 0.179551 | 41.415357 | 25.484325 | 10.372226 | 5.600155 | 8.090038 | 1541.197008 | 78.807802 | 46.451613 | 6634.715089 |
| min | 0.000000 | 0.140228 | 0.000000 | 1899.000000 | 1969.000000 | 32.647300 | -27.069900 | 0.007907 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 2.800000 | 0.356400 | 2004.000000 | 1971.000000 | 2018.000000 | 46.861100 | 5.029700 | 90.338261 | 0.000000 | 0.000000 | 0.000000 |
| 50% | 8.200000 | 0.385600 | 2012.000000 | 1997.000000 | 2021.000000 | 50.207635 | 9.255472 | 792.753846 | 0.000000 | 0.000000 | 0.000000 |
| 75% | 25.000000 | 0.580939 | 2017.000000 | 2009.000000 | 2023.000000 | 52.630700 | 12.850190 | 2021.408119 | 0.000000 | 0.000000 | 0.000000 |
| max | 6000.000000 | 0.917460 | 2030.000000 | 2020.000000 | 2051.000000 | 71.012300 | 39.655350 | 16840.000000 | 9500.000000 | 1800.000000 | 421000.000000 |
Sometimes, we also want to store a DataFrame for later use. There are many different file formats tabular data can be stored in, including HTML, JSON, Excel, Parquet, Feather, etc. Here, let’s say we want to store the DataFrame as CSV (comma-separated values) file under the name “tmp.csv”.
df.to_csv("tmp.csv")
df
| Name | Fueltype | Technology | Set | Country | Capacity | Efficiency | DateIn | DateRetrofit | DateOut | lat | lon | Duration | Volume_Mm3 | DamHeight_m | StorageCapacity_MWh | EIC | projectID | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 0 | Kernkraftwerk Emsland | Nuclear | Steam Turbine | PP | Germany | 1336.000 | 0.33 | 1988.0 | 1988.0 | 2023.0 | 52.472897 | 7.324140 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'MASTR': {'MASTR-SEE944567587799'}, 'ENTSOE':... |
| 1 | Brokdorf | Nuclear | Steam Turbine | PP | Germany | 1410.000 | 0.33 | 1986.0 | 1986.0 | 2021.0 | 53.850830 | 9.344720 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'MASTR': {'MASTR-SEE951462745445'}, 'ENTSOE':... |
| 2 | Borssele | Hard Coal | Steam Turbine | PP | Netherlands | 485.000 | NaN | 1973.0 | NaN | 2034.0 | 51.433200 | 3.716000 | NaN | 0.0 | 0.0 | 0.0 | {'49W000000000054X'} | {'BEYONDCOAL': {'BEYOND-NL-2'}, 'ENTSOE': {'49... |
| 3 | Gemeinschaftskernkraftwerk Neckarwestheim | Nuclear | Steam Turbine | PP | Germany | 1310.000 | 0.33 | 1976.0 | 1989.0 | 2023.0 | 49.040019 | 9.176408 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'MASTR': {'MASTR-SEE985577062814'}, 'ENTSOE':... |
| 4 | Isar | Nuclear | Steam Turbine | PP | Germany | 1410.000 | 0.33 | 1979.0 | 1988.0 | 2023.0 | 48.605600 | 12.293150 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'MASTR': {'MASTR-SEE943690268513'}, 'ENTSOE':... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 29858 | Zf Frd Prufstand | Oil | NaN | PP | Germany | 6.166 | NaN | 2000.0 | NaN | NaN | 47.666864 | 9.486685 | NaN | 0.0 | 0.0 | 0.0 | {nan, nan, nan, nan} | {'MASTR': {'MASTR-SEE937736737958', 'MASTR-SEE... |
| 29859 | Zf Pas Bhkw | Natural Gas | NaN | CHP | Germany | 1.182 | NaN | 2017.0 | NaN | NaN | 48.603866 | 13.422240 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'MASTR': {'MASTR-SEE954959703560'}} |
| 29860 | Zf Sbr Prufstand | Oil | NaN | PP | Germany | 2.400 | NaN | 2007.0 | NaN | NaN | 49.217758 | 6.973464 | NaN | 0.0 | 0.0 | 0.0 | {nan, nan} | {'MASTR': {'MASTR-SEE948501471533', 'MASTR-SEE... |
| 29861 | Zi | Other | NaN | CHP | Germany | 47.590 | NaN | 1980.0 | NaN | NaN | 52.299865 | 11.672539 | NaN | 0.0 | 0.0 | 0.0 | {nan, nan} | {'MASTR': {'MASTR-SEE950355589975', 'MASTR-SEE... |
| 29862 | Zre | Waste | NaN | PP | Germany | 18.800 | NaN | 2026.0 | NaN | NaN | 53.583875 | 9.911636 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'MASTR': {'MASTR-SEE995521057735'}} |
29565 rows × 18 columns
Grouping and Aggregation#
Both Series and DataFrame objects have a groupby method, which allows you to group and aggregate the data based on the values of one or more columns.
It accepts a variety of arguments, but the simplest way to think about it is that you pass another series, whose unique values are used to split the original object into different groups.
Here’s an example which retrieves the total generation capacity per country.
grouped = df.groupby("Country").Capacity.sum()
grouped.sort_values(ascending=False).head(10)
Country
Germany 266203.749352
Spain 150108.858726
United Kingdom 149679.992061
France 146501.108342
Italy 101488.386461
Ukraine 59477.000000
Poland 54328.702513
Sweden 48139.886625
Netherlands 43014.294952
Norway 39809.690000
Name: Capacity, dtype: float64
Such chaining of multiple operations is very common with pandas.
Let’s break apart this operation a bit. The workflow with groupby can be divided into three general steps:
Split: Partition the data into different groups based on some criterion.
Apply: Do some calculation (e.g. aggregation or transformation) within each group.
Combine: Put the results back together into a single object.

Grouping is not only possible on a single columns, but also on multiple columns. For instance,
we might want to group the capacities by country and fuel type. To achieve this, we pass a list of functions to the groupby functions.
capacities = df.groupby(["Country", "Fueltype"]).Capacity.sum()
capacities
Country Fueltype
Albania Hydro 1743.9
Other 98.0
Solar 294.5
Wind 234.0
Austria Hard Coal 1331.4
...
United Kingdom Other 55.0
Solar 11668.6
Solid Biomass 4919.0
Waste 288.9
Wind 38670.3
Name: Capacity, Length: 242, dtype: float64
By grouping by multiple attributes, our index becomes a pd.MultiIndex (a hierarchical index with multiple levels.
capacities.index[:5]
MultiIndex([('Albania', 'Hydro'),
('Albania', 'Other'),
('Albania', 'Solar'),
('Albania', 'Wind'),
('Austria', 'Hard Coal')],
names=['Country', 'Fueltype'])
type(capacities.index)
pandas.core.indexes.multi.MultiIndex
df.nsmallest(10, "Capacity")
| Name | Fueltype | Technology | Set | Country | Capacity | Efficiency | DateIn | DateRetrofit | DateOut | lat | lon | Duration | Volume_Mm3 | DamHeight_m | StorageCapacity_MWh | EIC | projectID | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 963 | Stve | Hydro | Run-Of-River | PP | United Kingdom | 0.0 | NaN | 1951.0 | 1951.0 | NaN | 56.014999 | -5.121000 | NaN | 0.0 | 0.0 | 0.0 | {'48WSTN00000STVEO'} | {'ENTSOE': {'48WSTN00000STVEO'}, 'JRC': {'JRC-... |
| 8853 | Banovici | Lignite | NaN | PP | Bosnia and Herzegovina | 0.0 | NaN | NaN | NaN | NaN | 44.400000 | 18.533300 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'BEYONDCOAL': {'BEYOND-BA-6'}} |
| 8854 | Bielefeld | Hard Coal | NaN | PP | Germany | 0.0 | NaN | 1980.0 | NaN | 2012.0 | 52.033500 | 8.538500 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'BEYONDCOAL': {'BEYOND-DE-25'}} |
| 8855 | Bugojno | Lignite | NaN | PP | Bosnia and Herzegovina | 0.0 | NaN | NaN | NaN | NaN | 44.050000 | 17.450000 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'BEYONDCOAL': {'BEYOND-BA-7'}} |
| 8857 | Compostilla | Lignite | NaN | PP | Spain | 0.0 | NaN | 1961.0 | NaN | 2020.0 | 42.627500 | -6.563300 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'BEYONDCOAL': {'BEYOND-ES-27'}} |
| 8858 | Datteln | Hard Coal | NaN | PP | Germany | 0.0 | NaN | 1964.0 | NaN | 2013.0 | 51.629300 | 7.330700 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'BEYONDCOAL': {'BEYOND-DE-50'}} |
| 8860 | Gacko | Lignite | NaN | PP | Bosnia and Herzegovina | 0.0 | NaN | NaN | NaN | NaN | 43.172259 | 18.512728 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'BEYONDCOAL': {'BEYOND-BA-8'}} |
| 8861 | Hafen Muenster | Hard Coal | NaN | PP | Germany | 0.0 | NaN | 1977.0 | NaN | 2005.0 | 51.950300 | 7.640500 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'BEYONDCOAL': {'BEYOND-DE-174'}} |
| 8863 | Kamengrad | Lignite | NaN | PP | Bosnia and Herzegovina | 0.0 | NaN | NaN | NaN | NaN | 44.766667 | 16.666667 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'BEYONDCOAL': {'BEYOND-BA-11'}} |
| 8864 | Kolubara | Lignite | NaN | PP | Serbia | 0.0 | NaN | NaN | NaN | NaN | 44.467500 | 20.284440 | NaN | 0.0 | 0.0 | 0.0 | {nan} | {'BEYONDCOAL': {'BEYOND-RS-7'}} |
We can use the .unstack function to reshape the multi-indexed pd.Series into a pd.DataFrame which has the second index level as columns.
capacities.unstack().fillna(0.0).T.round(1).to_csv(
"capacities_by_country_and_fueltype.csv"
)
In summary, the typical workflow with pandas consists of reading data from files, inspecting and cleaning the data, performing analysis through transformation and aggregation, visualizing the results, and storing the processed data for later use.
Exercises#
Power Plants Data#
In this exercise, we will use the powerplants.csv dataset from the powerplantmatching project. This dataset contains information about various power plants, including their names, countries, fuel types, capacities, and more.
URL: https://raw.githubusercontent.com/PyPSA/powerplantmatching/master/powerplants.csv
Task 1: Load the dataset into a pandas DataFrame.
Task 2: Run the function .describe() on the DataFrame.
Task 3: Provide a list of unique fuel types and technologies included in the dataset.
Note
Look in the pandas documentation for functions that might be useful to solve these tasks.
Task 4: Filter the dataset by power plants with the fuel type “Hard Coal”.
Task 5: Identify the 5 largest coal power plants. In which countries are they located? When were they built?
Task 6: Identify the power plant with the longest name.
Task 7: Identify the 10 northernmost powerplants. What type of power plants are they?
Task 8: What is the average start year of each fuel type? Sort the fuel types by their average start year in ascending order and round to the nearest integer.
Wind and Solar Capacity Factors#
In this exercise, we will work with a time series dataset containing hourly wind and solar capacity factors for Ireland, taken from model.energy.
Task 1: Use pd.read_csv to load the dataset from the following URL into a pandas DataFrame. Ensure that the time stamps are treated as pd.DatetimeIndex.
Task 2: Calculate the mean capacity factor for wind and solar over the entire time period.
Task 3: Calculate the correlation between wind and solar capacity factors.
Note
Go to the pandas documentation for functions that might be useful to solve these tasks.
Task 4: Plot the wind and solar capacity factors for the month of May.
Task 5: Plot the weekly average capacity factors for wind and solar over the entire time period.
Task 6: Go to model.energy and retrieve the time series for another region of your choice. Recreate the analysis above and compare the results.
Note
Look for “Download Comma-Separated-Variable (CSV) file of data” in Step 2.

